A novel metagenome-derived viral RNA polymerase
25 October 2022, by Dr. Yuchen Han
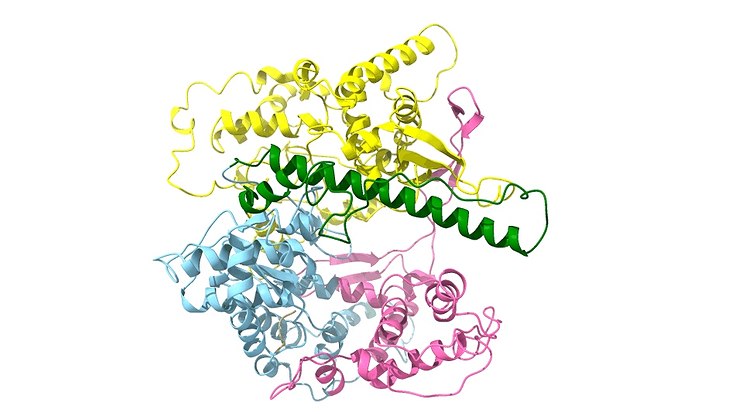
Photo: UHH/Mikrobiologie
The mining of genomes from non-cultivated microorganisms using metagenomics is a powerful tool to discover novel proteins and other valuable biomolecules. However, function-based metagenome searches are often limited by the time-consuming expression of the active proteins in various heterologous host systems. We here report the initial characterization of novel single-subunit bacteriophage RNA polymerase, EM1 RNAP, identified from a metagenome data set obtained from an elephant dung microbiome. EM1 RNAP and its promoter sequence are distantly related to T7 RNA polymerase. Using EM1 RNAP and a translation-competent Escherichia coli extract, we have developed an efficient medium-throughput pipeline and protocol allowing the expression of metagenome-derived genes and the production of proteins in cell-free system is sufficient for the initial testing of the predicted activities. Here, we have successfully identified and verified 12 enzymes acting on bis(2-hydroxyethyl) terephthalate (BHET) in a completely clone-free approach and proposed an in vitro high-throughput metagenomic screening method.
Published on 25 October 2022 in Scientific Reports
The protocol of the metagenomic screening via in vitro expression system can be found in the book Metagenomics: Methods in Molecular Biology (vol 2555. Humana, New York).
For the interest on EM1 RNAP, please contact to our IP management by click here.

